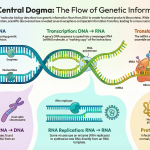
Introduction: Preparing the Blueprint for Life

Imagine a highly organized construction crew arriving at a site. Before a single foundation can be poured, they must survey the land, mark the exact starting point, get the right permits, and move the heavy machinery into position. Only after this meticulous preparation can the actual building begin.

In a remarkably similar way, our cells prepare their own blueprint for life—our DNA—before copying it. Before a cell can divide, it must create a perfect duplicate of its entire genome. This requires an intricate preparation phase where specialized protein machinery is assembled at precise locations on the DNA. This crucial setup process is known as pre-replicative complex (pre-RC) assembly.

This document will walk you through this assembly process step-by-step. We will introduce the key protein “actors” and explain their specific roles in this elegant and essential biological sequence.

Stage 1: Initiation of Replication
Now that we’ve set the scene, let’s identify the specific location where all this important work takes place.
1. The “Construction Site”: The Origin of Replication
In the vast landscape of a chromosome, replication doesn’t just start anywhere. It begins at specific, designated locations called origins of replication. These sites act as the official starting points for the entire process of DNA duplication.
Think of an origin of replication as a “start here” marker on a map or a designated launchpad for a mission. It is the specific site where the construction crew—the protein machinery—initiates the assembly of the pre-RC. While some eukaryotes, like yeast, have origins defined by a specific DNA sequence, the system in more complex organisms like humans is different. Instead of recognizing a simple sequence, the machinery appears to identify origins based on epigenetic marks or the specific 3D structure of the chromatin, such as particular modifications to the nucleosomes in the origin region.
Regardless of the exact method of identification, the origin is the fundamental “construction site” where the process of preparing for DNA replication begins.
With the construction site located, it’s time to meet the specialized crew responsible for the job.
2. The Cast of Characters: Meet the Key Protein Actors
The assembly of the pre-RC is a biological play with a precise cast of protein complexes. Each has a specialized role to perform in a specific order. Let’s meet the four main actors.
| Protein Actor | Primary Role | Simple Analogy |
| Origin Recognition Complex (ORC) | Acts as the “initiator” that recognizes and binds to the origin of replication to kick off the entire process. | The Site Surveyor: Finds the exact spot on the map and plants the first flag. |
| Cdc6 | An ATP-binding protein that is recruited by ORC and is required for loading the MCM helicase onto the DNA. | The Permit Officer: Arrives after the surveyor and authorizes the delivery of heavy machinery to the site. |
| Cdt1 | A key “licensing factor” that works with Cdc6 to recruit and load the MCM helicase onto the DNA. | The Gatekeeper: Works alongside the Permit Officer to physically open the gate and guide the machinery into place. |
| MCM2-7 Helicase | A six-protein complex that is the future DNA helicase; its main job will be to unwind the DNA strands. Its loading “licenses” the origin for replication. | The Unzipping Machine: The critical piece of heavy machinery that, once loaded, is ready to separate the two DNA strands. |

Now that you’ve met the cast, let’s see how they work together in a precise sequence to prepare the DNA.
3. The Assembly Line: A Step-by-Step Guide to Pre-RC Formation
Now that we’ve met the cast, let’s examine the precise choreography of their actions. The assembly of the pre-RC is not random; it’s a meticulously ordered sequence, like an assembly line, that occurs during the G1 phase of the cell cycle—a period of growth when the cellular environment is properly configured for this critical preparation.

1. Step 1: ORC Plants the Flag. The Origin Recognition Complex (ORC) is the first actor on the scene. It identifies the specific origin of replication site on the DNA and binds directly to it. This foundational step requires energy in the form of ATP and marks the location for all subsequent activity.
2. Step 2: The Loading Crew Arrives. Once ORC is anchored to the origin, it serves as a docking station, recruiting Cdc6 and Cdt1. These two proteins, sometimes called “licensing factors,” are both essential for the next step. They associate with the ORC-bound DNA independently of one another but must work together to bring in the main piece of machinery.
3. Step 3: The Unzipping Machine is Loaded. This is the central event of pre-RC assembly. With the help of Cdc6 and Cdt1, two ring-shaped MCM2-7 helicase complexes are loaded onto the DNA. These two hexamers (six-protein rings) are arranged in a head-to-head fashion, forming a double hexamer that completely encircles the double-stranded DNA, poised and ready to begin unwinding once it is activated in S phase.

4. Step 4: The Origin is “Licensed” and Ready. The successful loading of the MCM double hexamer completes the pre-RC. At this point, the origin is officially “licensed”—it is now fully prepared and approved to begin DNA replication once the cell transitions into the S phase. After their loading job is complete, ORC and Cdc6 can be removed from the chromatin, leaving the MCM complex poised for action.

This carefully choreographed assembly is complete, but why does the cell go to all this trouble?
4. The Importance of Order: Preventing Catastrophic Errors
The brilliance of this entire process lies in a simple but powerful strategy: the cell strictly separates when it can build the replication machinery from when it can turn it on. This temporal separation is the ultimate safeguard against catastrophic errors. By separating the permission to replicate (licensing in G1) from the act of replication (firing in S phase), the cell enforces a non-negotiable rule: the genome must be copied exactly once per cell cycle.
If an origin were allowed to “fire” more than once, a cell would end up with extra copies of certain genes. This phenomenon, known as re-replication, is extremely dangerous and can lead to genomic instability and disease. To prevent this, the cell employs redundant safety mechanisms.
• Cyclin-Dependent Kinases (CDKs): These master regulatory proteins have a crucial bipartite role. Their activity levels rise and fall with the cell cycle, creating a “two-for-one” control switch.
◦ Pre-RC assembly (licensing) is only permitted when CDK activity is low, which occurs during the G1 phase.
◦ As the cell enters S phase, CDK activity becomes high. This same rise in CDK activity both triggers the firing of the already-assembled pre-RCs to start replication and, at the same time, prevents the formation of any new pre-RCs. This ensures origins cannot be licensed again until the next cell cycle.

• Geminin: This is another inhibitory protein that provides an additional layer of security.
◦ Geminin levels are low in G1 but accumulate as the cell enters S phase.
◦ Its specific job is to bind to and block Cdt1, one of the essential loading factors. By inhibiting the “gatekeeper,” geminin effectively shuts down any illicit attempts to load a new MCM helicase onto the DNA after replication has already begun.
With that final piece of insight, you now have a complete picture of this foundational process.
The Stage is Set
Revisiting our analogy, the construction site has been meticulously prepared: the land has been surveyed and marked (ORC), the permits have been approved (Cdc6 and Cdt1), and the heavy machinery is assembled and ready to go (MCM helicase). With the stage now set, the cell is ready for the next act: the incredible feat of copying its entire genetic blueprint—an elegant process fundamental to the continuity of all eukaryotic life.
Replication After Launch: Eukaryotic DNA Elongation and Termination
Eukaryotic DNA replication is a highly orchestrated process, occurring in three primary stages: initiation, elongation, and termination. Once the initiation phase successfully assembles the pre-replicative complex (pre-RC) and activates the necessary machinery, the cell transitions into the critical phase of DNA synthesis: elongation. This stage involves coordinated synthesis across multiple replication forks until the entire genome is duplicated, culminating in termination.
Stage 2: Elongation—Building the New Strands
Elongation involves unwinding the parental DNA and synthesizing two new daughter strands simultaneously, a complex challenge due to the antiparallel nature of DNA.
1. The Core Replication Machinery
The replication machinery begins with the conversion of the inert pre-RC into an active replisome, driven by Dbf4-dependent kinase (DDK) and cyclin-dependent kinase (CDK) activity. The central unwinding engine is the CMG helicase (Cdc45–Mcm2-7–GINS complex), which separates the DNA duplex into single strands.
As the DNA strands separate, they form Y-shaped structures called replication forks. To prevent the separated single-stranded DNA (ssDNA) from re-annealing or forming secondary structures, it is coated by Replication Protein A (RPA).

2. Synthesis Direction and Strand Differentiation
DNA polymerase enzymes can only add new nucleotides in the 5′ to 3′ direction. Because the two parental strands run antiparallel, this results in two distinct synthesis mechanisms:

• The Leading Strand: This strand is oriented such that synthesis proceeds continuously toward the advancing replication fork as the helicase unwinds the double-stranded DNA. It requires only a single RNA primer to initiate replication. The bulk of continuous synthesis on the leading strand is performed by DNA polymerase ε (Pol ε).
• The Lagging Strand: This strand must be synthesized away from the replication fork. Since DNA polymerase can only add nucleotides in the 5′ to 3′ direction, the lagging strand is synthesized in short pieces called Okazaki fragments.

3. Priming and Polymerization
DNA polymerase cannot initiate a new strand from scratch; it requires an existing strand onto which it can add nucleotides. This precursor strand, or primer, is supplied by primase, a specialized RNA polymerase that creates a short RNA polynucleotide strand complementary to the DNA template.
• On both strands, the short RNA primer is then extended by the DNA polymerase α (Pol α), which adds a short stretch (10 to 20 bases) of deoxyribonucleotides.
• Once this initial segment is laid down, Pol α is replaced by the main replicative polymerases: Pol ε for the leading strand and DNA polymerase δ (Pol δ) for the lagging strand.
• To ensure high speed and continuous synthesis (processivity), the sliding clamp protein PCNA (Proliferating Cell Nuclear Antigen) encircles the DNA and tethers the polymerases. PCNA is loaded onto the primed template junctions by the Replication Factor C (RFC) complex.
4. Lagging Strand Maturation (Cleanup)
The synthesis of the lagging strand is followed by an extensive cleanup process necessary to produce a continuous DNA molecule.
• Primer Displacement and Removal: As Pol δ synthesizes a new Okazaki fragment, it eventually encounters the RNA primer of the previously synthesized fragment, displacing it along with a small segment of DNA to create a flap.
• The ribonucleotides composing the primers must be removed and replaced with DNA nucleotides. This is achieved by cellular enzymes, specifically RNase H, which digests the RNA part hybridized to DNA, and FEN1 (Flap Endonuclease 1), which removes the resulting flaps.
• A free-floating DNA polymerase fills the resulting gap with DNA nucleotides. This action leaves breaks in the sugar-phosphate backbone called nicks.
• Ligation: In the final step of DNA replication, the enzyme ligase (specifically DNA ligase I) joins the sugar-phosphate backbones at each nick site, making the new strand continuous and completing the daughter DNA molecule.

Stage 3: Termination—Finishing the Job
The elongation stage concludes with the termination of replication across the chromosome and the resolution of linear ends.

1. Fork Convergence and CMG Disassembly
Termination generally occurs when two replication forks collide or converge. A replication fork terminates when the leading strand of one replication bubble meets the lagging strand of the adjacent bubble, or when the lagging strand runs into the 5′ end of a previous Okazaki fragment.
The convergence of two CMG helicase complexes at the end of DNA synthesis triggers the disassembly of the replication machinery. This disassembly requires the SCF E3 ubiquitin ligase complex to ubiquitinate the MCM protein (MCM7 subunit). This poly-ubiquitinated MCM complex is then instantly recognized by the segregase protein p97, which catalyzes the disassembly of the helicase complex from the DNA strand, thus terminating replication.
2. The End Replication Problem
Due to the fundamental requirement for a primer and the inability of DNA polymerase to initiate synthesis at the very end of a linear chromosome, the lagging strand synthesis leaves a short single-stranded DNA overhang (ssDNA) on the parental strand. If this were left unresolved, each daughter chromosome would progressively shorten with every cell division, known as the end replication problem.
Eukaryotic cells resolve this issue using specialized telomere regions (repetitive sequences at chromosome ends) and the enzyme telomerase.
• Telomerase is a ribonucleoprotein that acts as a specialized DNA polymerase. It consists of a protein subunit called TERT (telomerase reverse transcriptase) and an RNA component.
• Telomerase uses its own RNA template to bind to and extend the 3′ end of the existing parental strand.
• This extension of the 3′ parental strand creates a longer template, allowing primase to lay down a final RNA primer and conventional lagging strand synthesis to fill the gap on the 5′ end of the daughter strand.
• The final overhang created by the telomerase-synthesized DNA (which is considered “junk DNA”) is removed by FEN1, thereby protecting the actual genomic DNA from being shortened.

Once the new DNA strand is continuous and the linear ends are protected, the daughter DNA molecule is complete.
Image Summary







Reference
Bell, S. P., & Dutta, A. (2002). DNA replication in eukaryotic cells. In Annual Review of Biochemistry (Vol. 71, pp. 333–374). https://doi.org/10.1146/annurev.biochem.71.110601.135425
Burgers, P. M. J., & Kunkel, T. A. (2017). Downloaded from www.annualreviews.org. Guest (guest). https://doi.org/10.1146/annurev-biochem
Costa, A., & Diffley, J. F. X. (2025). The Initiation of Eukaryotic DNA Replication. 47, 15. https://doi.org/10.1146/annurev-biochem-072321
Hu, Y., & Stillman, B. (2023). Origins of DNA replication in eukaryotes. In Molecular Cell (Vol. 83, Issue 3, pp. 352–372). Cell Press. https://doi.org/10.1016/j.molcel.2022.12.024
Masai, H., Matsumoto, S., You, Z., Yoshizawa-Sugata, N., & Oda, M. (2010). Eukaryotic chromosome DNA replication: Where, when, and how? In Annual Review of Biochemistry (Vol. 79, pp. 89–130). https://doi.org/10.1146/annurev.biochem.052308.103205
Méchali, M. (2010). Eukaryotic DNA replication origins: Many choices for appropriate answers. In Nature Reviews Molecular Cell Biology (Vol. 11, Issue 10, pp. 728–738). Nature Publishing Group. https://doi.org/10.1038/nrm2976
Parker, M. W., Botchan, M. R., & Berger, J. M. (2017). Mechanisms and regulation of DNA replication initiation in eukaryotes. In Critical Reviews in Biochemistry and Molecular Biology (Vol. 52, Issue 2, pp. 107–144). Taylor and Francis Ltd. https://doi.org/10.1080/10409238.2016.1274717
Stillman, B., Diffley, J. F. X., & Iwasa, J. H. (2025). Mechanisms for licensing origins of DNA replication in eukaryotic cells. In Nature Structural and Molecular Biology (Vol. 32, Issue 7, pp. 1143–1153). Nature Research. https://doi.org/10.1038/s41594-025-01587-5
https://en.wikipedia.org/wiki/Eukaryotic_DNA_replication
https://bio.libretexts.org/Bookshelves/Microbiology/Microbiology_(Boundless)/07%3A_Microbial_Genetics/7.03%3A_DNA_Replication/7.3B%3A_DNA_Replication_in_Eukaryotes
https://bio.libretexts.org/Bookshelves/Introductory_and_General_Biology/General_Biology_(Boundless)/14%3A_DNA_Structure_and_Function/14.03%3A_DNA_Replication/14.3C%3A_DNA_Replication_in_Eukaryotes