Introduction: The Cell’s Protein Factory
Imagine a high-tech 3D printer that reads a digital blueprint, one line of code at a time, to build an intricate and functional object. The process of translation in a bacterial cell is remarkably similar. It’s the fundamental process where the cell reads the genetic instructions encoded in a molecule called messenger RNA (mRNA) to build proteins—the essential machines and structures of life.
This complex assembly is not a chaotic scramble but a highly organized, three-act play:
1. Initiation: Setting up the machinery.
2. Elongation: Building the protein chain, piece by piece.
3. Termination: Finishing the job and releasing the final product.
Let’s explore how bacteria flawlessly execute this essential task.
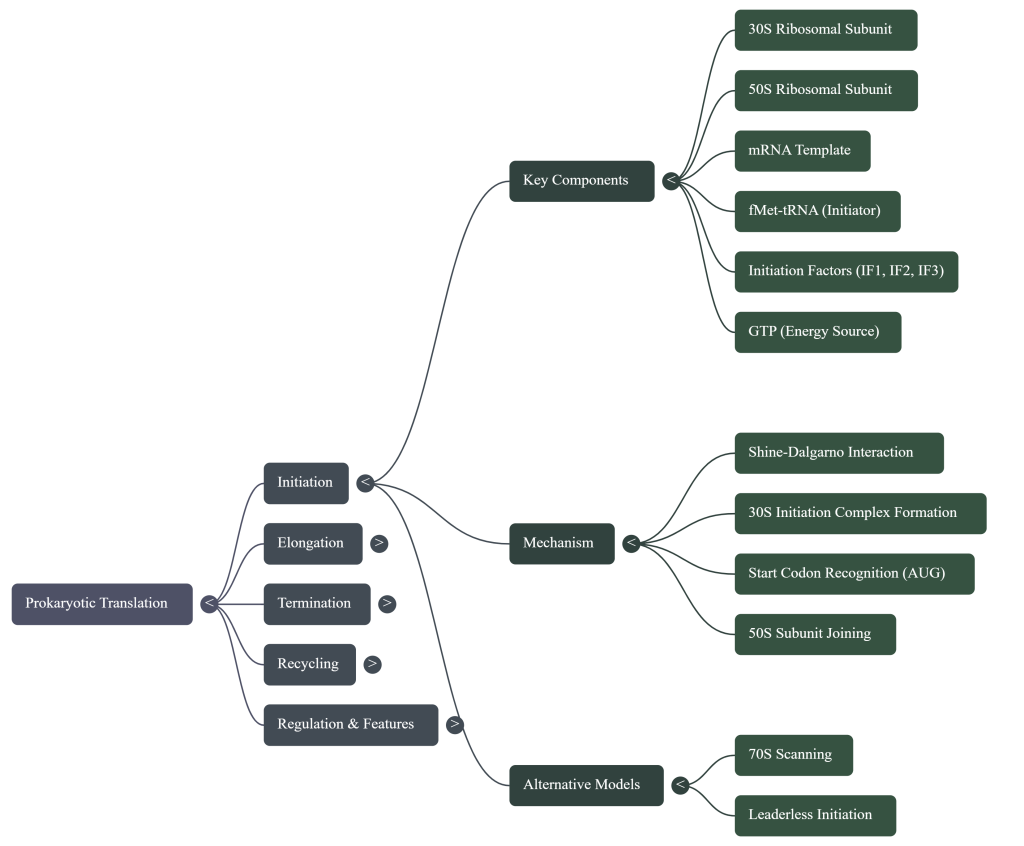
1. The Cast of Characters: Meet the Molecular Machinery
Before diving into the process, it’s crucial to meet the key players involved in this molecular construction project. Each component has a specific and vital role.
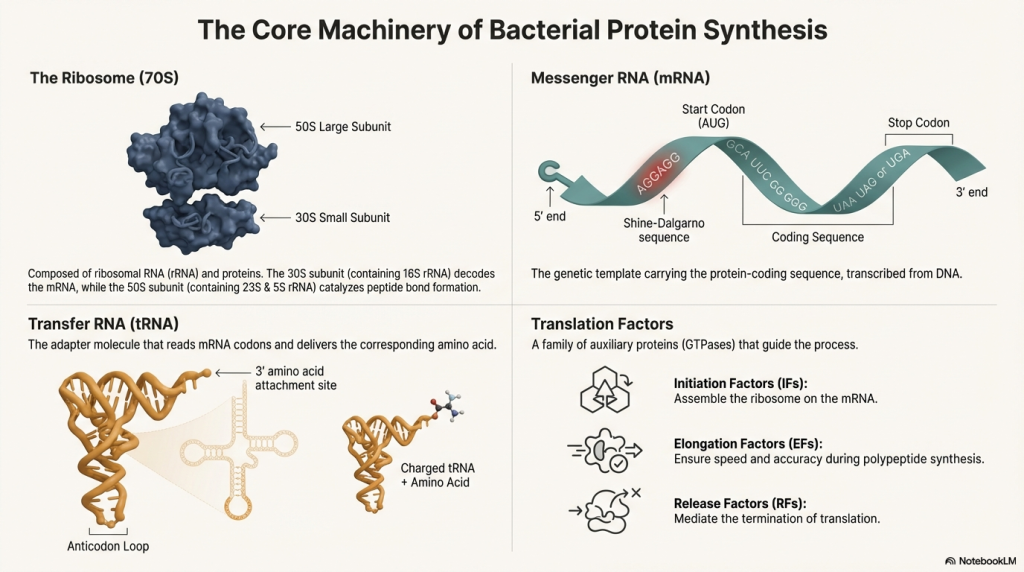
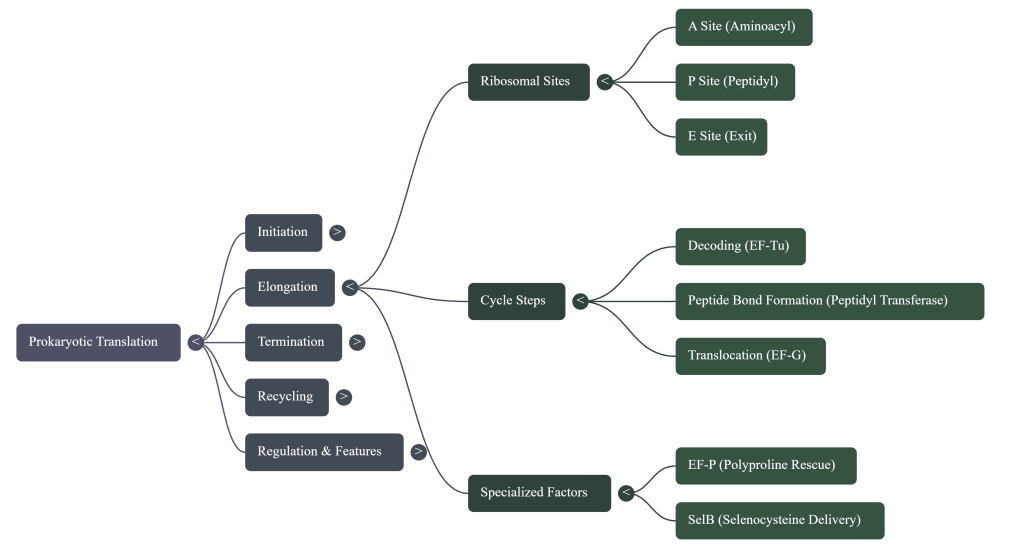
• The Ribosome (The Factory) This is the cell’s protein-building machine. In bacteria, it’s composed of two parts: a small 30S subunit and a large 50S subunit that join together to form a functional 70S ribosome. (A quick note for curious students: the “S” stands for Svedberg, a unit of sedimentation rate, not mass, which is why 30 + 50 doesn’t equal 70!). It features three crucial worksites, much like an assembly line:
◦ A site (Arrival): The entry point where new, charged tRNA molecules arrive.
◦ P site (Peptide-building): The workstation where the growing protein chain is held.
◦ E site (Exit): The exit door for empty tRNA molecules after they’ve dropped off their amino acid.
• Messenger RNA (mRNA) (The Blueprint) The mRNA is the instruction manual, a direct copy of a gene from the cell’s DNA. Its genetic language is written in three-letter “words” called codons. The mRNA blueprint contains three critical signals for the ribosome:
◦ Shine-Dalgarno Sequence: A “Dock Here” sign upstream of the start codon. By ensuring the ribosome locks on at the perfect spot, this sequence establishes the correct reading frame, preventing the production of a garbled, non-functional protein.
◦ AUG Start Codon: The signal that tells the ribosome to “Begin Building.”
◦ Stop Codons (UAA, UAG, UGA): The “End of the Line” signal that halts the process.
• Transfer RNA (tRNA) (The Delivery Trucks) Each tRNA is a specialized delivery truck responsible for carrying one specific type of amino acid. Every tRNA has two key features: an anticodon, which is a three-letter sequence that reads the mRNA codon, and an attachment site for its specific amino acid. A special initiator tRNA carries the very first amino acid in every bacterial protein, N-formyl-methionine (fMet).
• Factors (The Supervisors) A group of helper proteins, called Initiation Factors (IF1, IF2, IF3), Elongation Factors (EF-Tu, EF-G), and Release Factors (RF1, RF2, RF3), act as supervisors. They are required to manage, assist, and ensure each stage of the translation process runs smoothly and accurately.
• GTP (The Energy Source) Guanosine triphosphate (GTP) is the energy currency that powers several key steps of translation, including the binding of new tRNAs and the movement of the ribosome along the mRNA.
Now that we’ve met the key players, let’s see how they work together in the first act of protein synthesis.
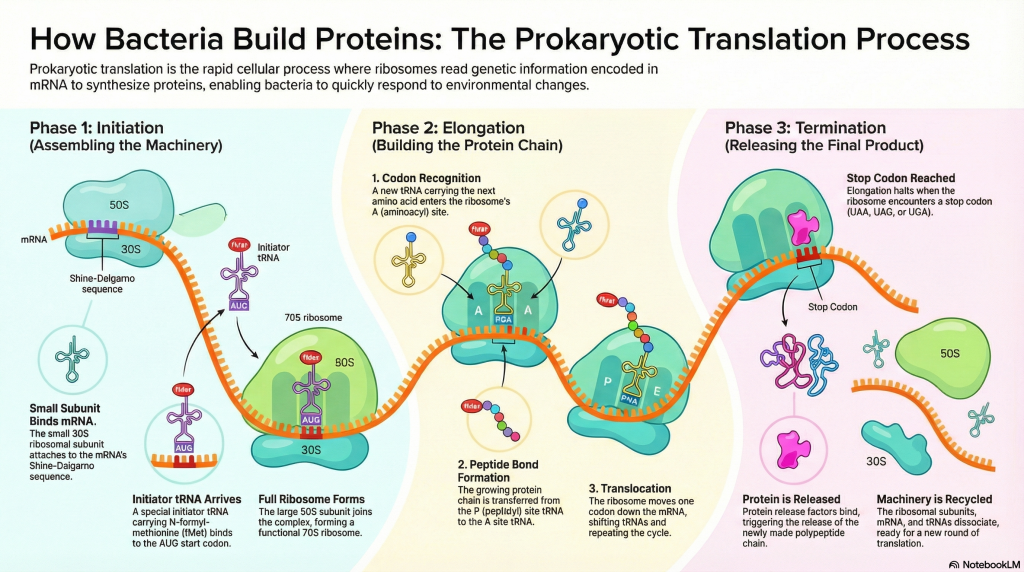
Act 1: Initiation – Setting Up the Factory
Initiation is all about assembling the ribosome at the correct starting point on the mRNA blueprint. This setup ensures that the protein is built from the right starting amino acid and that the genetic code is read in the correct frame.
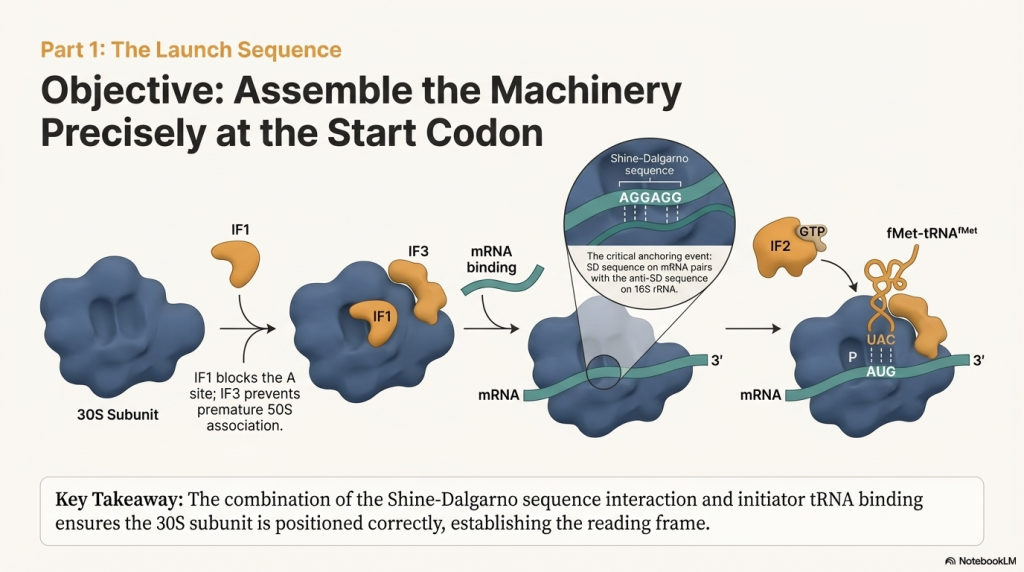
1. The Small Subunit Binds the mRNA The process begins when the small 30S ribosomal subunit, assisted by initiation factors (IF1 and IF3), locates and binds to the mRNA. It doesn’t bind just anywhere; it recognizes the Shine-Dalgarno sequence. This interaction occurs when the Shine-Dalgarno sequence on the mRNA base-pairs with a complementary sequence on the 16S rRNA, a component of the 30S subunit. This anchors the subunit and positions it perfectly so that the AUG start codon is situated in what will become the P site.
2. The First tRNA Arrives Next, the special initiator tRNA, carrying N-formyl-methionine (fMet), is escorted to the small subunit by another initiation factor, IF2. The anticodon of this initiator tRNA base-pairs with the AUG start codon, locking the first building block into place in the P site. While fMet begins every bacterial protein, it is usually removed after translation is complete.
3. The Full Ribosome Assembles With the mRNA and initiator tRNA in position, the large 50S subunit joins the complex. This locks all the pieces together, forming a complete and functional 70S ribosome. This final assembly step triggers the release of all the initiation factors, leaving the factory ready for production with a free A site, poised to accept the next amino acid.
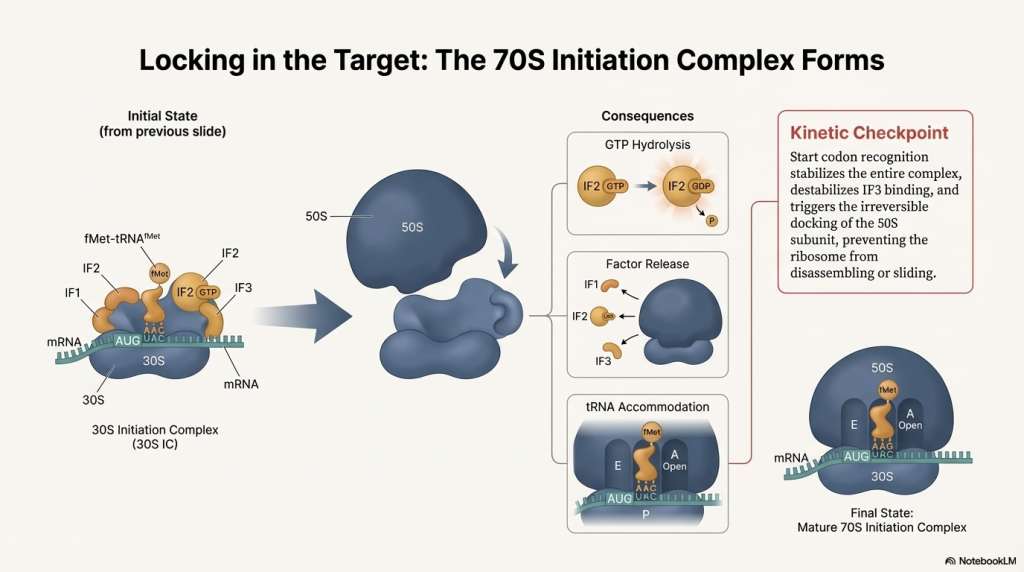
With the factory fully assembled and the first building block in place, the assembly line is ready to start running at full speed.
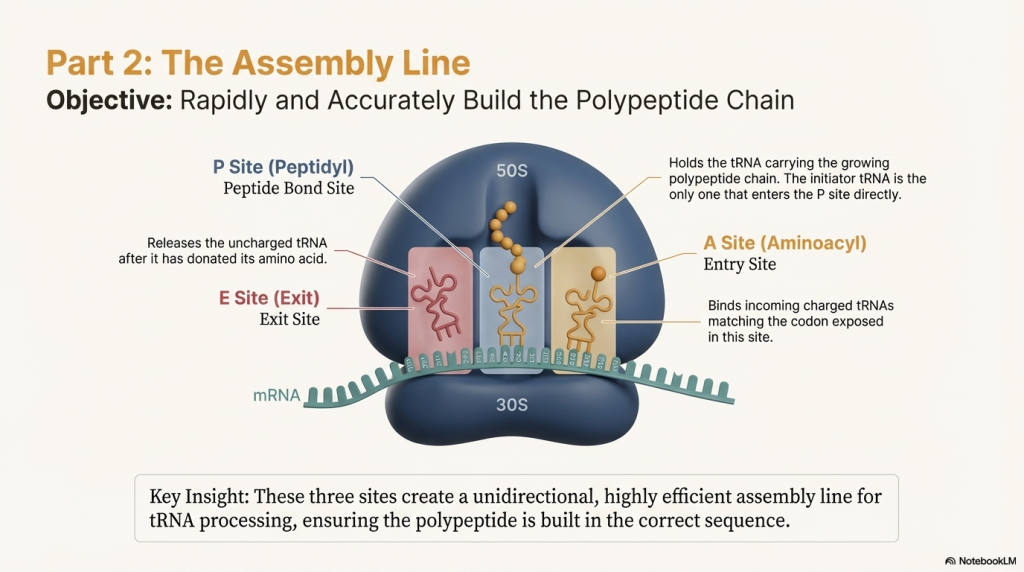
Act 2: Elongation – The Assembly Line in Action
Elongation is a highly efficient, repeating cycle where the protein chain is built one amino acid at a time. Think of it as the ribosome’s assembly line, moving steadily down the mRNA blueprint.
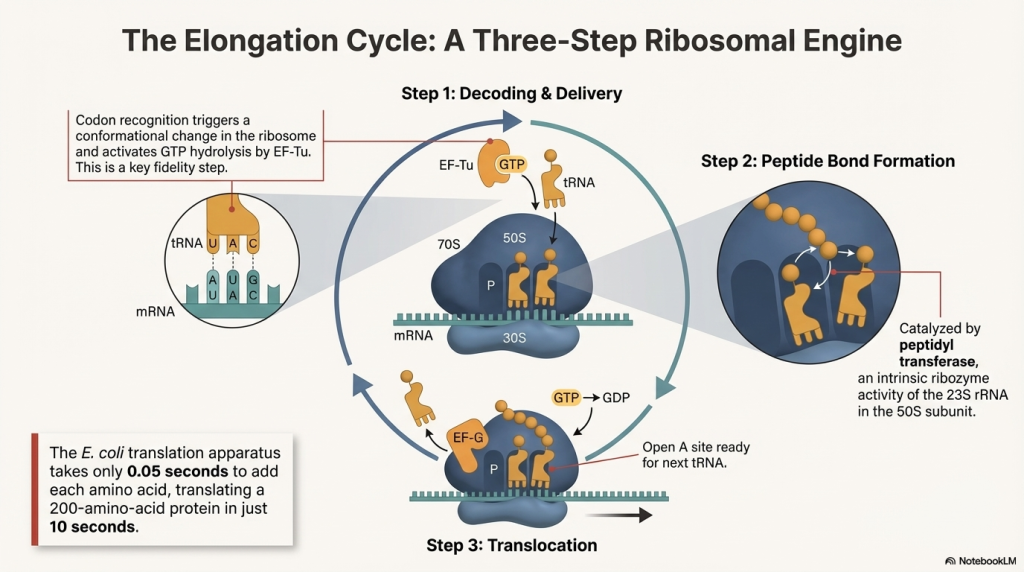
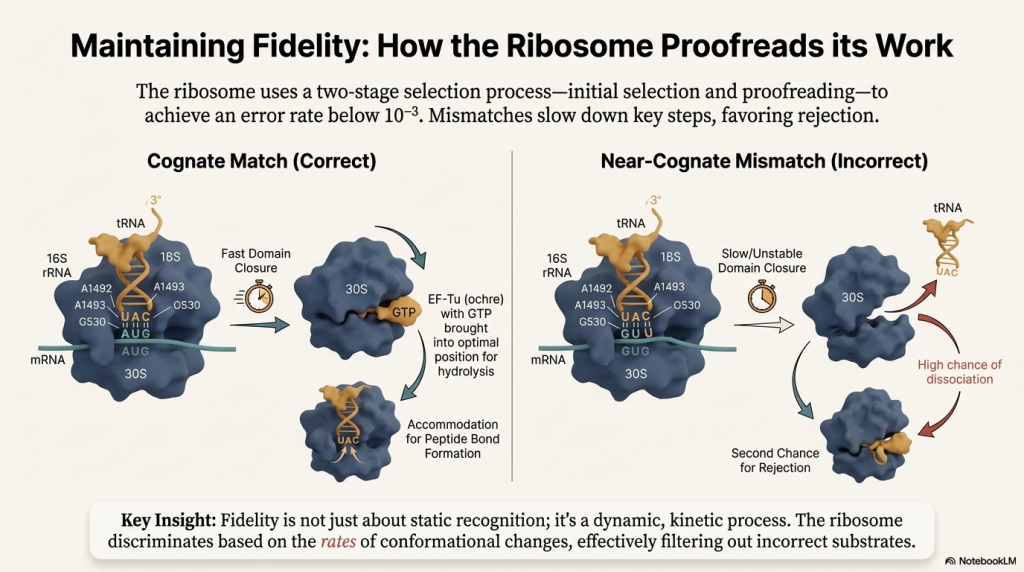
1. A New tRNA Arrives (Codon Recognition) An elongation factor called EF-Tu, powered by GTP, escorts a new charged tRNA into the empty A site. The ribosome performs a crucial proofreading step: only the tRNA whose anticodon is a perfect match for the mRNA codon in the A site will bind. Incorrect tRNAs are rejected.
2. A Peptide Bond is Formed (Peptide Transfer) The ribosome’s own catalytic machinery springs into action. This catalytic activity, called peptidyl transferase, is not performed by a protein but by the 23S rRNA of the large 50S subunit—making the ribosome a true ribozyme (an RNA-based enzyme). It catalyzes the formation of a peptide bond, linking the amino acid from the P-site tRNA onto the amino acid on the A-site tRNA. This action transfers the entire growing polypeptide chain to the tRNA in the A site.
3. The Ribosome Moves (Translocation) Powered by another elongation factor, EF-G, and the hydrolysis of GTP, the entire ribosome shifts exactly one codon (three bases) down the mRNA. This movement has three immediate consequences:
◦ The now-empty tRNA that was in the P site moves to the E site and is ejected from the ribosome.
◦ The tRNA holding the growing protein chain, which was in the A site, moves into the P site.
◦ The A site is now empty again, ready to accept the next tRNA delivery truck.
4. Repeat This three-step cycle of arrival, peptide bond formation, and translocation repeats for each codon on the mRNA. This process is remarkably fast, synthesizing proteins at a rate of about 18 amino acids per second. To put that speed in context, it’s much slower than DNA replication, where bacterial machinery can add 1,000 nucleotides per second, reflecting the greater complexity of polymerizing 20 different types of amino acids.
This highly efficient cycle continues, rapidly extending the protein chain until a special signal on the blueprint indicates that the job is done.
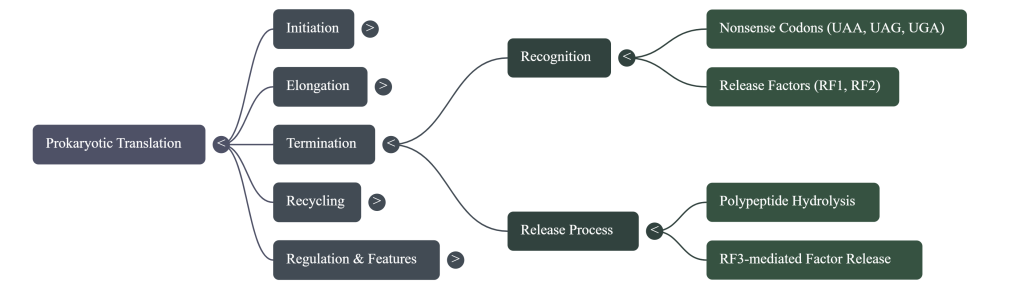
Act 3: Termination – Finishing the Job
Termination occurs when the ribosome reaches the end of the protein-coding sequence on the mRNA. This final phase ensures the newly built protein is released and the machinery is disassembled for reuse.
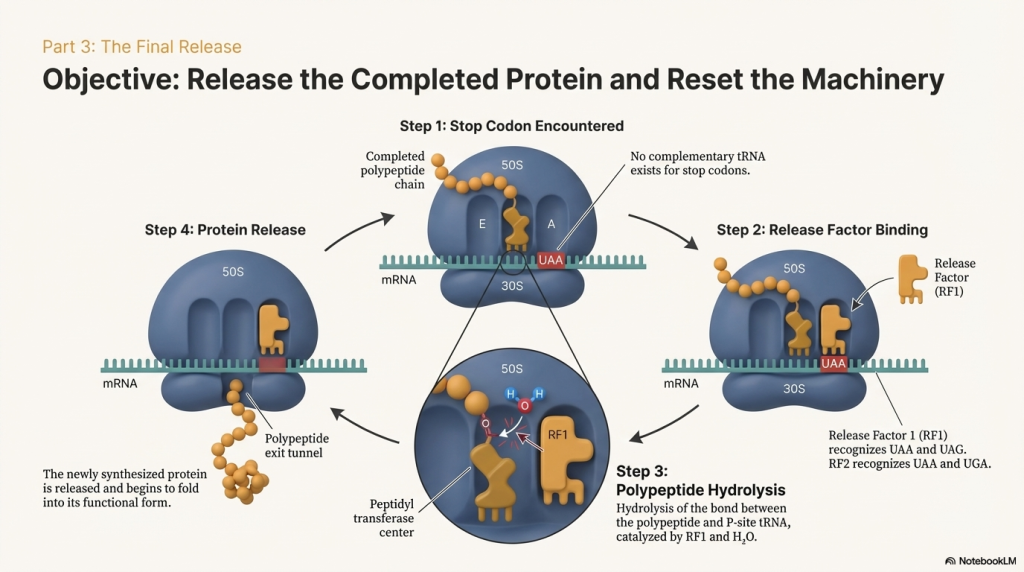
• Stop Codon Recognition The elongation cycle halts when the ribosome’s A site aligns with one of the three stop codons: UAA, UAG, or UGA. There are no tRNAs that recognize these codons.
• Release Factor Binding Instead of a tRNA, a protein called a release factor (RF1 recognizes UAA and UAG; RF2 recognizes UAA and UGA) binds to the stop codon in the A site.
• Protein Release The binding of the release factor causes the ribosome’s peptidyl transferase center to use a water molecule instead of an amino acid. This water molecule hydrolyzes the ester bond connecting the completed polypeptide to the tRNA in the P site, releasing the new protein from the ribosome.
• Disassembly After the protein is freed, another factor, RF3, catalyzes the release of RF1 and RF2 from the ribosome. The entire translation complex then falls apart. The ribosomal subunits, the mRNA, and the remaining tRNA dissociate from each other. These components are now free to be recycled and start a new round of translation on another mRNA molecule.
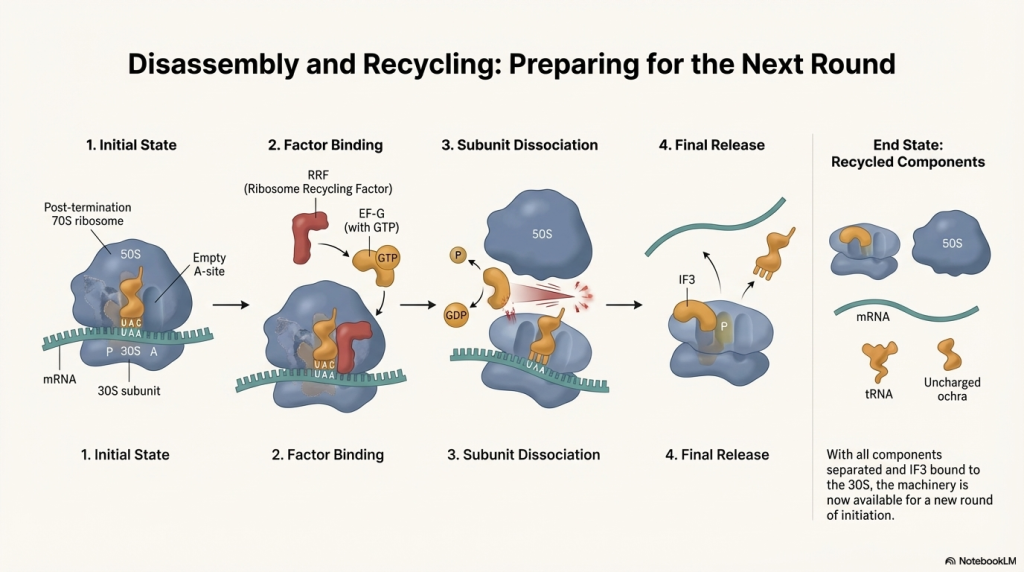
The protein is now complete and released to do its job, and the cellular machinery is reset for the next protein-building assignment.
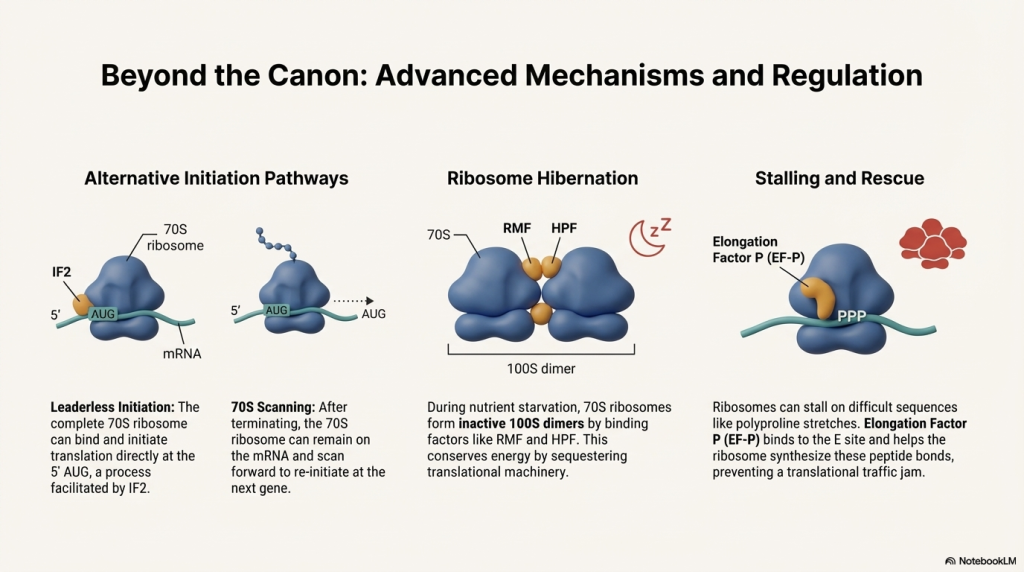
Summary and Key Distinctions
This table provides a high-level overview of the entire process for quick review.
| Phase | What Happens | Key Players |
| Initiation | The ribosome assembles on the mRNA at the start codon. | 30S & 50S subunits, mRNA, Shine-Dalgarno sequence, Initiator fMet-tRNA, Initiation Factors (IF1, IF2, IF3), GTP |
| Elongation | Amino acids are added one by one to a growing chain as the ribosome moves down the mRNA. | Charged tRNAs, Elongation Factors (EF-Tu, EF-G), GTP, Peptidyl transferase (23S rRNA) |
| Termination | The ribosome reaches a stop codon, the new protein is released, and the machinery disassembles. | Stop codons (UAA, UAG, UGA), Release Factors (RF1, RF2, RF3) |
A defining feature of prokaryotic life is its efficiency. One of the most important distinctions between bacteria and more complex eukaryotes is how they handle transcription and translation.
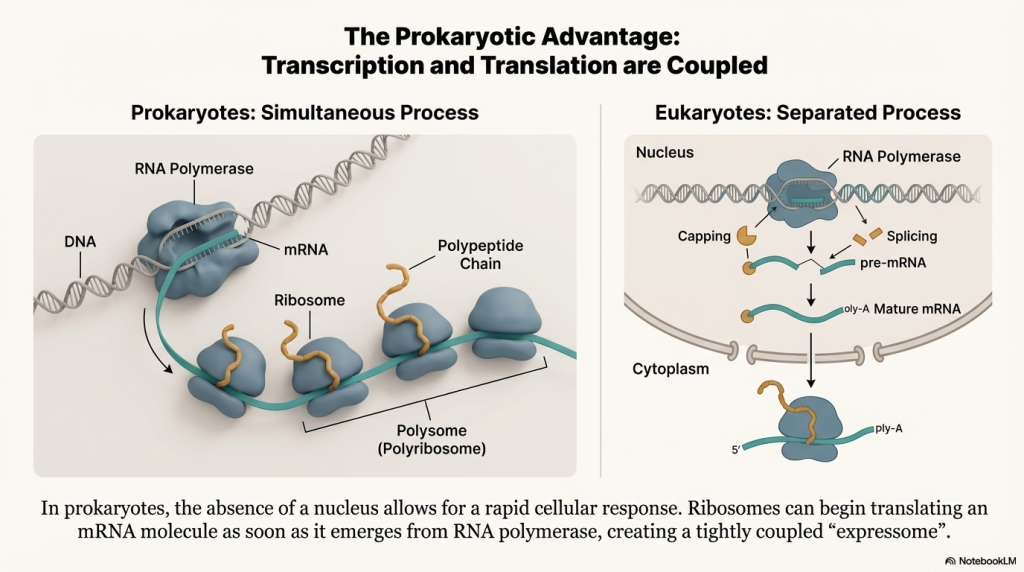
| Feature | Prokaryotes (e.g., E. coli) |
| Location & Timing | Transcription and translation happen at the same time in the cytoplasm. The ribosome can attach to the mRNA while it’s still being made. |
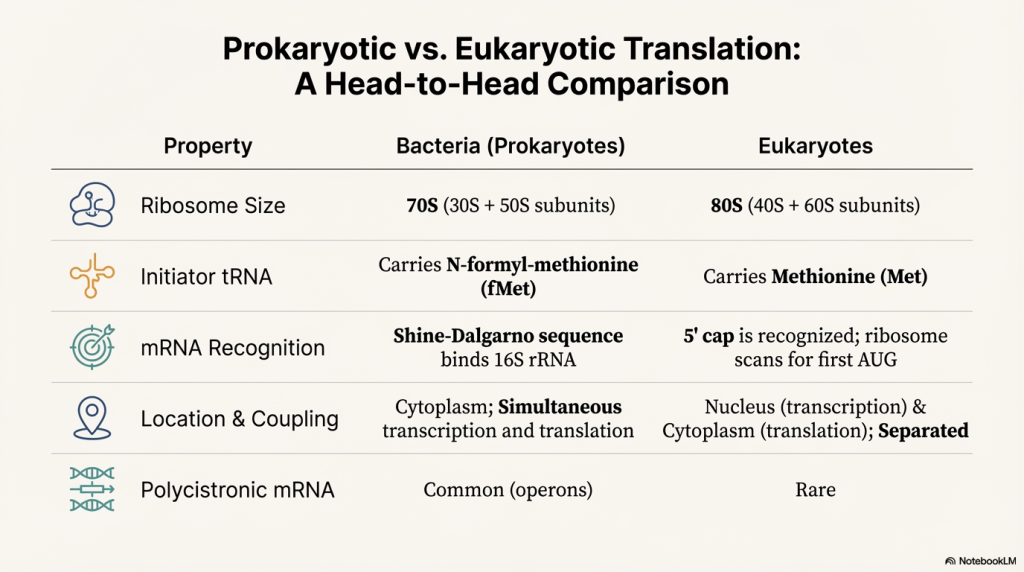
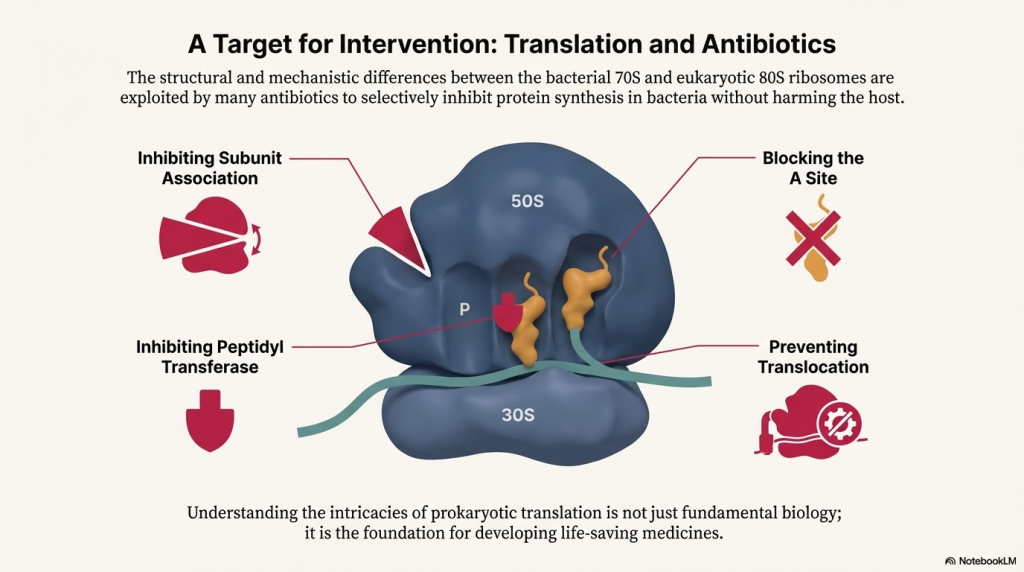
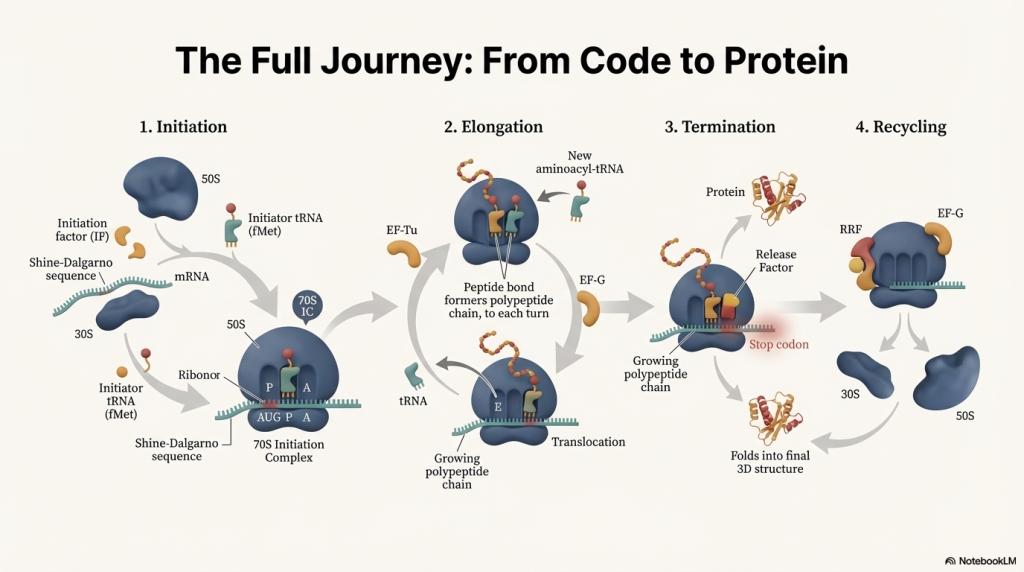
Image Summary
References
Rodnina MV. Translation in Prokaryotes. Cold Spring Harb Perspect Biol. 2018 Sep 4;10(9):a032664. doi: 10.1101/cshperspect.a032664. PMID: 29661790; PMCID: PMC6120702.
https://en.wikipedia.org/wiki/Bacterial_translation
https://courses.lumenlearning.com/wm-biology1/chapter/prokaryotic-translation/
https://bio.libretexts.org/Courses/Lumen_Learning/Biology_for_Majors_I_(Lumen)/12%3A_Module_10-_DNA_Transcription_and_Translation/12.15%3A_Prokaryotic_Translation
https://www.biologydiscussion.com/cell/prokaryotes/translation-in-prokaryotes-genetics/38022